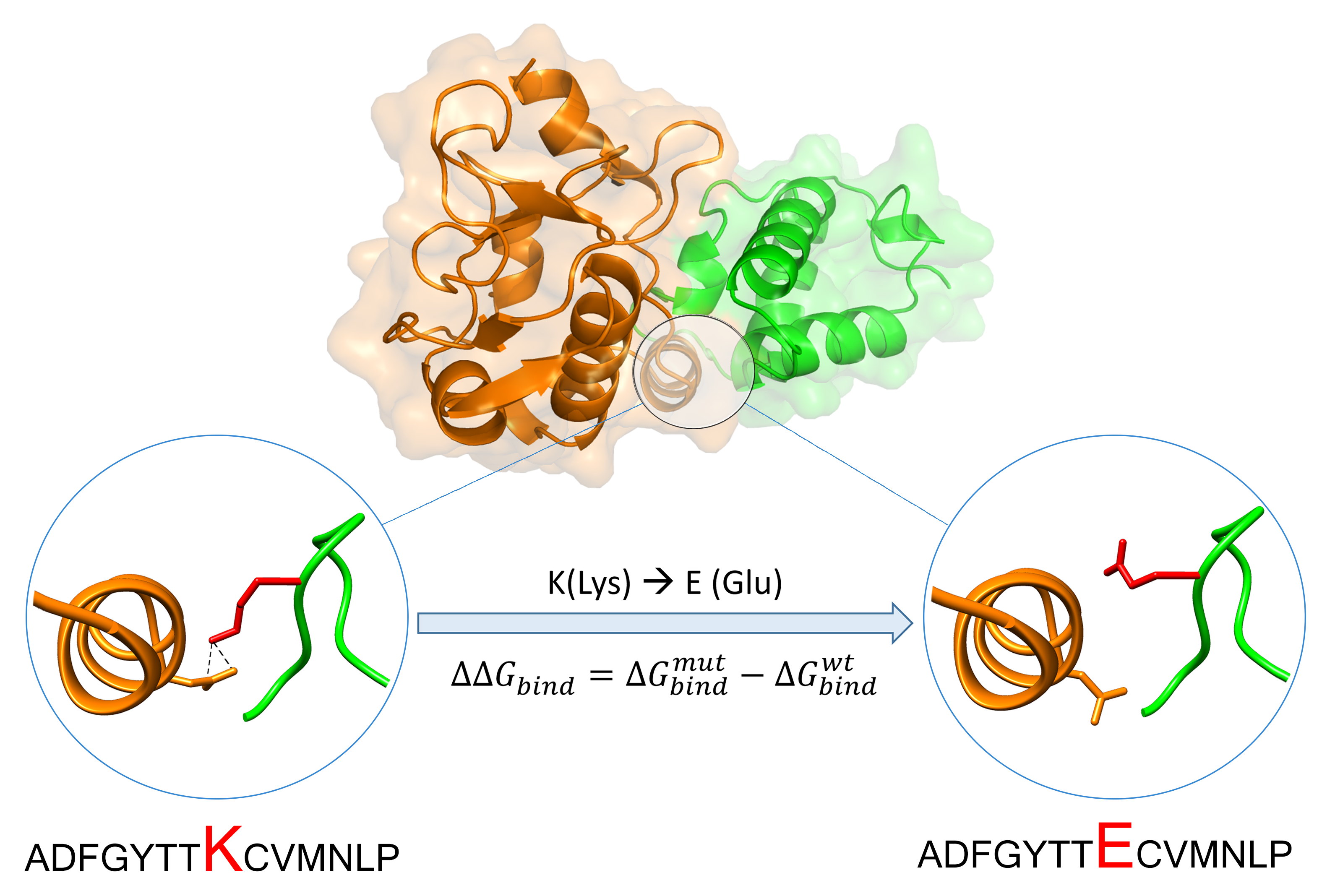
MutaBind and MutaBind2
MutaBind and MutaBind2 evaluate the effects of sequence variants and disease mutations on protein interactions and calculates the quantitative changes in binding affinity. It uses molecular mechanics force fields, statistical potentials and fast side-chain optimization algorithms. The server maps mutations on a structural protein complex, calculates the associated changes in binding affinity, determines the deleterious effect of a mutation, estimates the confidence of this prediction and produces a mutant structural model for download. MutaBind can be applied to a large number of problems, including determination of potential driver mutations in cancer and other diseases, elucidation of the effects of sequence variants on protein fitness in evolution and protein design. Use the server